About
Hello! I am Vamsi Nallapareddy! I am currently working as a Research Assistant at the Orengo Laboratory under the supervision of Prof. Christine Orengo at University College London.
I worked on multiple projects during the course of my bachelor’s degree in different domains such as Computational Biology, Deep Learning, Computer Vision, Natural Language Processing, and Reinforcement Learning. Exploring these different fields has helped me find a passion for the application of deep learning techniques to learn more from biological data in order to build applications and tools that can make a positive impact in the world.
Apart from my academic work, I like to read books, design, and write.
I am actively looking for Ph.D. opportunities starting in the Fall of 2022. Please feel free to get in touch with me if you want to have a chat!
- Computational Biology
- Bioinformatics
- Deep Learning
B.E. in Computer Science, 2021
BITS Pilani - Hyderabad
Experience
Supervisor: Prof. Christine Orengo
Projects:
- CATHe: Remote homologue detection using embeddings from protein langauge models [In collaboration with Dr. Sameer Velankar’s Lab, EMBL-EBI & Prof. Burkhard Rost’s Lab, Technical University Munich]
- Study of TriTerperne Synthase (TTS) enzymes [In collaboration with Prof. Janet Thornton’s Lab, EMBL-EBI & Prof. Anne Osbourn’s Lab, the John Innes Institute]
Supervisor: Prof. Christine Orengo
Projects:
- Deep learning based classification of protein sequences into CATH Structurally Similar Groups (SSGs) [In Collaboration with Prof. Burkhard Rost’s Lab, Technical University Munich]
- Protein melting point prediction using deep learning [In collaboration with Prof. Florian Hollfelder’s Lab, Cambridge University]
Supervisor: Prof. Min Xu
Project: Studying Computer Vision techniques to classify biological molecules in a Cryo-ET tomogram
Supervisor: Prof. Arne Elofsson
Project: Protein inter-residue distance prediction using features derived from Multiple Sequence Alignments (MSA)
Supervisor: Prof. Peter Peer
Project: Designing a deep learning based model to diagnose COVID-19 from Chest X-Ray (CXR) data
Awards
- Awarded the university merit based scholarship for being in the top 1% of my cohort.
- Recipient of the prestigious National Talent Search Examination (NTSE) scholarship provided by the Government of India
- This scholarship provides a monthly stipend to the recipient till the end of their education.
Projects
Adaptive Bitrate Estimation using Reinforcement Learning
Advanced the state-of-the-art A3C model implemented in Pensieve by increasing exploration using the Follow then Forage technique
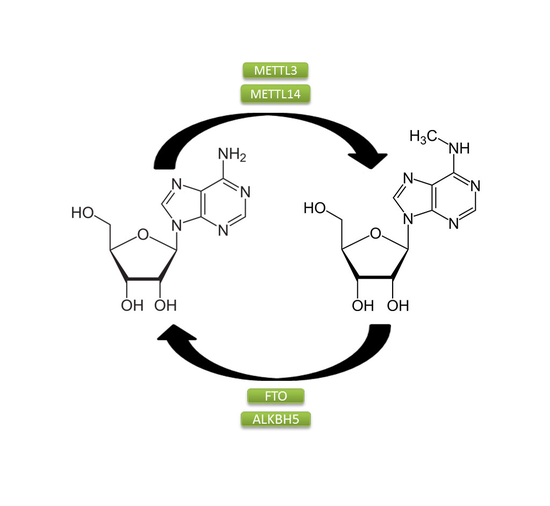
m6A Site Prediction
Trained a deep learning model to identify N6-MethylAdenosine (m6A) modifications in an RNA sequence
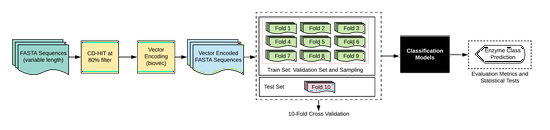
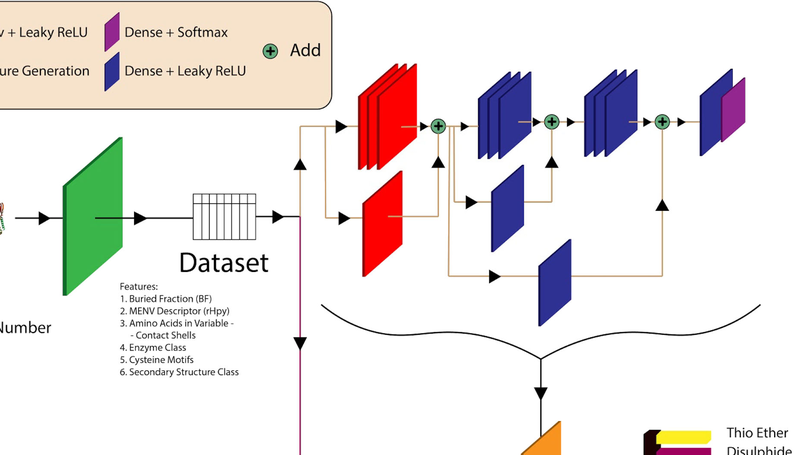
ABLE
An Attention Based Learning method for Enzyme Classification using protein sequences
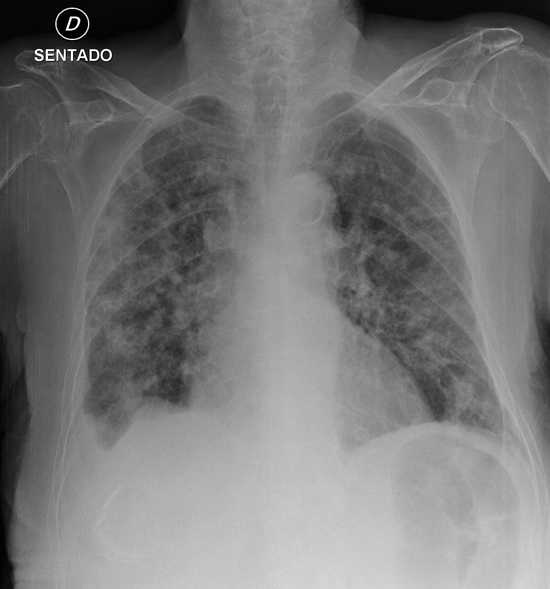
COVID-19 Diagnosis
Designed an deep learning model to diagnose COVID-19 from Chest X-rays
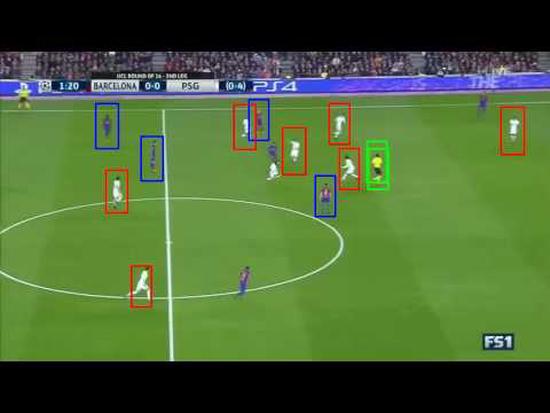
Player Detection in Football Match Videos
Constructed a deep learning model to detect, classify, and track players in a football match video

Tomogram Analysis
Classified sub-tomograms according to the biomolecules present in them using various deep learning techniques
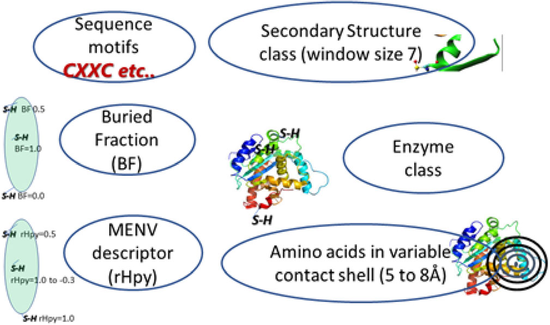
DeepCys
Deep learning based system to predict four different cysteine post-translational modifications
Serine Repeat Antigen 5 (SERA5) and Egress
Conducted protein-protein docking simulations to understand the role of SERA5 and SERA6 in the process of egress, pertinent to the malaria parasite Plasmodium falciparum
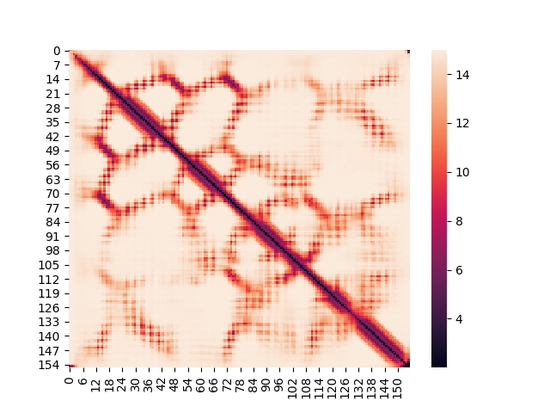
Inter-Residue Distance Prediction
Designed multiple deep learning models to predict the pair-wise distances for all the residues in a protein